Merge develop into branch
Showing
- .gitlab-ci.yml 23 additions, 17 deletions.gitlab-ci.yml
- README.md 1 addition, 1 deletionREADME.md
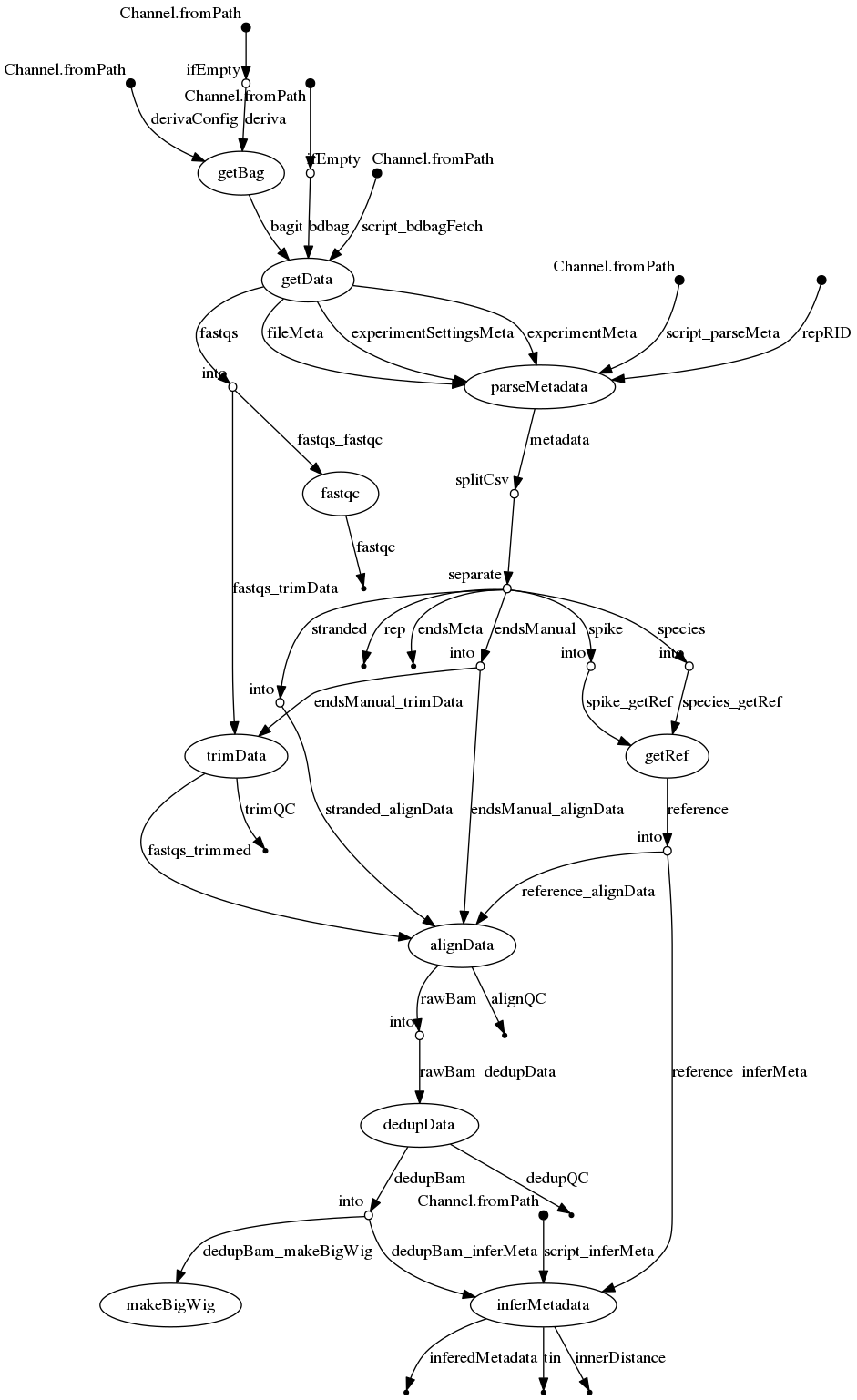
- docs/dag.png 0 additions, 0 deletionsdocs/dag.png
- workflow/conf/biohpc.config 1 addition, 1 deletionworkflow/conf/biohpc.config
- workflow/rna-seq.nf 19 additions, 13 deletionsworkflow/rna-seq.nf
- workflow/tests/test_dedupReads.py 8 additions, 2 deletionsworkflow/tests/test_dedupReads.py
| W: | H:
| W: | H:



