get 1kb center of enhancer ans figures.
Showing
- ES_D0_gro-seq_enhancers.bed 1304 additions, 0 deletionsES_D0_gro-seq_enhancers.bed
- ES_D0_gro-seq_enhancers_1kb.bed 1304 additions, 0 deletionsES_D0_gro-seq_enhancers_1kb.bed
- ES_D10_gro-seq_enhancers.bed 1099 additions, 0 deletionsES_D10_gro-seq_enhancers.bed
- ES_D10_gro-seq_enhancers_1kb.bed 1099 additions, 0 deletionsES_D10_gro-seq_enhancers_1kb.bed
- ES_D2_gro-seq_enhancers.bed 1535 additions, 0 deletionsES_D2_gro-seq_enhancers.bed
- ES_D2_gro-seq_enhancers_1kb.bed 1535 additions, 0 deletionsES_D2_gro-seq_enhancers_1kb.bed
- ES_D5_gro-seq_enhancers.bed 651 additions, 0 deletionsES_D5_gro-seq_enhancers.bed
- ES_D5_gro-seq_enhancers_1kb.bed 651 additions, 0 deletionsES_D5_gro-seq_enhancers_1kb.bed
- ES_D7_gro-seq_enhancers.bed 525 additions, 0 deletionsES_D7_gro-seq_enhancers.bed
- ES_D7_gro-seq_enhancers_1kb.bed 525 additions, 0 deletionsES_D7_gro-seq_enhancers_1kb.bed
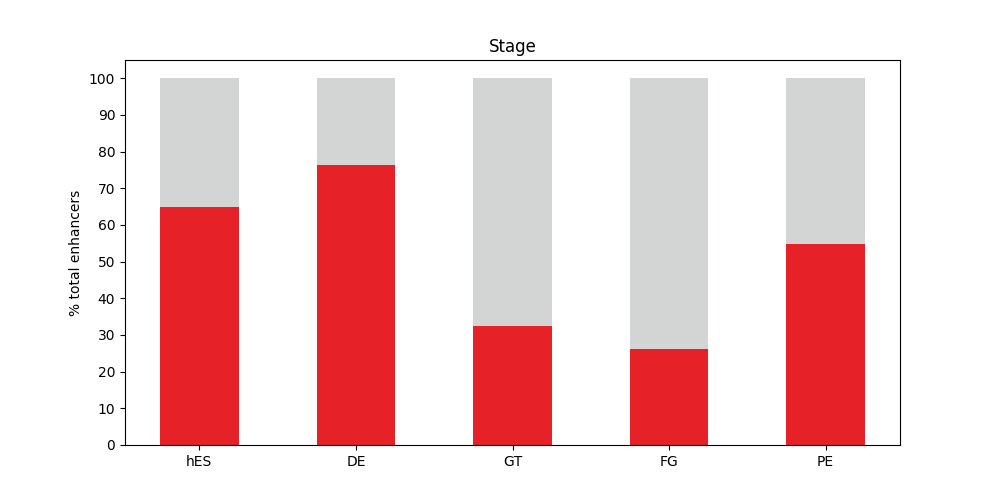
- Gro_percentages.png 0 additions, 0 deletionsGro_percentages.png
- SSP_filtered_peaks_merged.bed 1314 additions, 458 deletionsSSP_filtered_peaks_merged.bed
- SSP_filtered_peaks_sum.tsv 1306 additions, 0 deletionsSSP_filtered_peaks_sum.tsv
- gro-seq_enhancer_plots.py 102 additions, 0 deletionsgro-seq_enhancer_plots.py
- groseq_processing.sh 33 additions, 4 deletionsgroseq_processing.sh
- histone_centered_processing.sh 69 additions, 0 deletionshistone_centered_processing.sh
ES_D0_gro-seq_enhancers.bed
0 → 100644
This diff is collapsed.
ES_D0_gro-seq_enhancers_1kb.bed
0 → 100644
This diff is collapsed.
ES_D10_gro-seq_enhancers.bed
0 → 100644
This diff is collapsed.
ES_D10_gro-seq_enhancers_1kb.bed
0 → 100644
This diff is collapsed.
ES_D2_gro-seq_enhancers.bed
0 → 100644
This diff is collapsed.
ES_D2_gro-seq_enhancers_1kb.bed
0 → 100644
This diff is collapsed.
ES_D5_gro-seq_enhancers.bed
0 → 100644
This diff is collapsed.
ES_D5_gro-seq_enhancers_1kb.bed
0 → 100644
This diff is collapsed.
ES_D7_gro-seq_enhancers.bed
0 → 100644
This diff is collapsed.
ES_D7_gro-seq_enhancers_1kb.bed
0 → 100644
This diff is collapsed.
Gro_percentages.png
0 → 100644
15.4 KiB
This diff is collapsed.
SSP_filtered_peaks_sum.tsv
0 → 100644
This diff is collapsed.
gro-seq_enhancer_plots.py
0 → 100644
This diff is collapsed.