Showing
- .gitlab-ci.yml 6 additions, 6 deletions.gitlab-ci.yml
- CHANGELOG.md 1 addition, 0 deletionsCHANGELOG.md
- README.md 4 additions, 1 deletionREADME.md
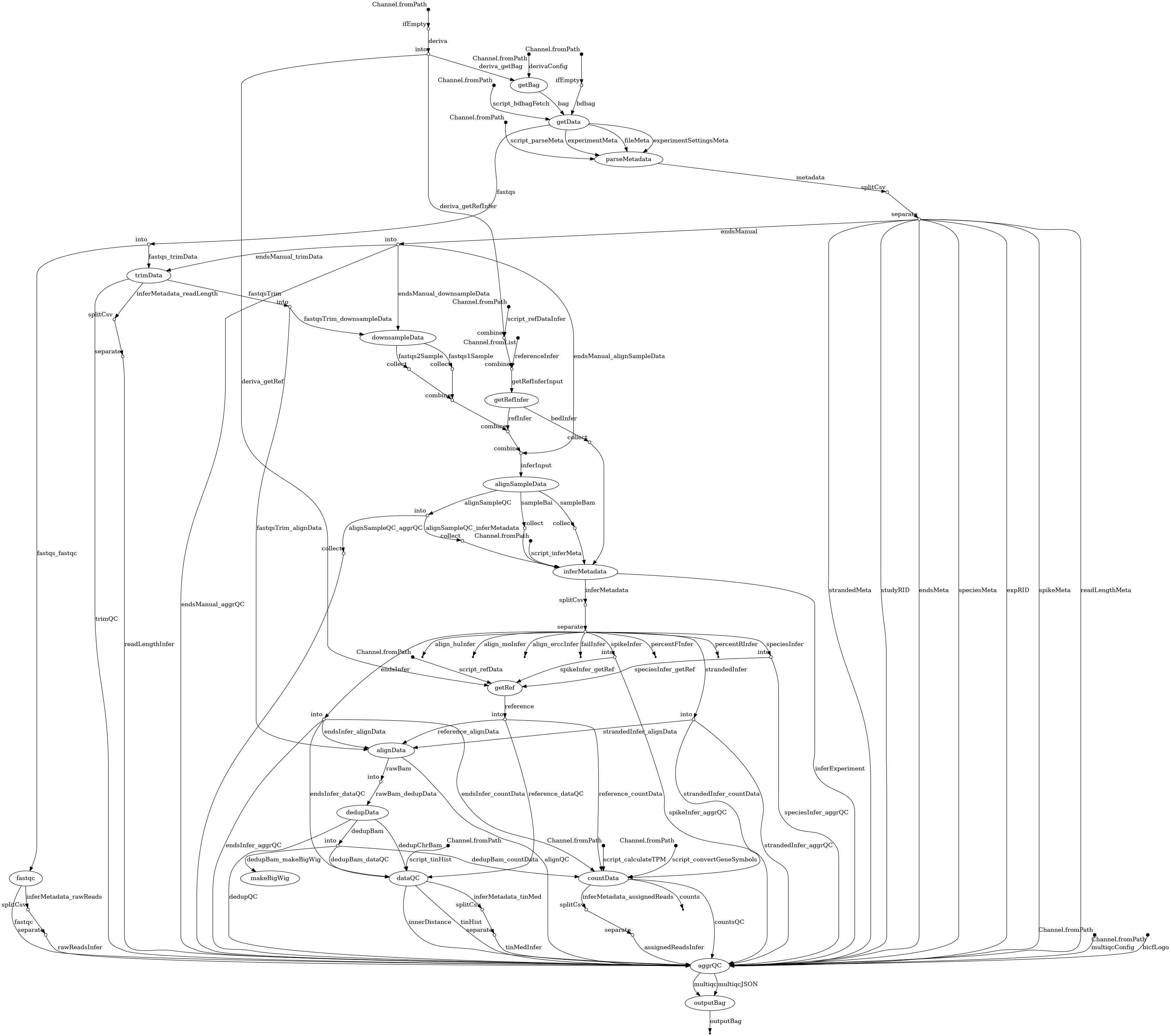
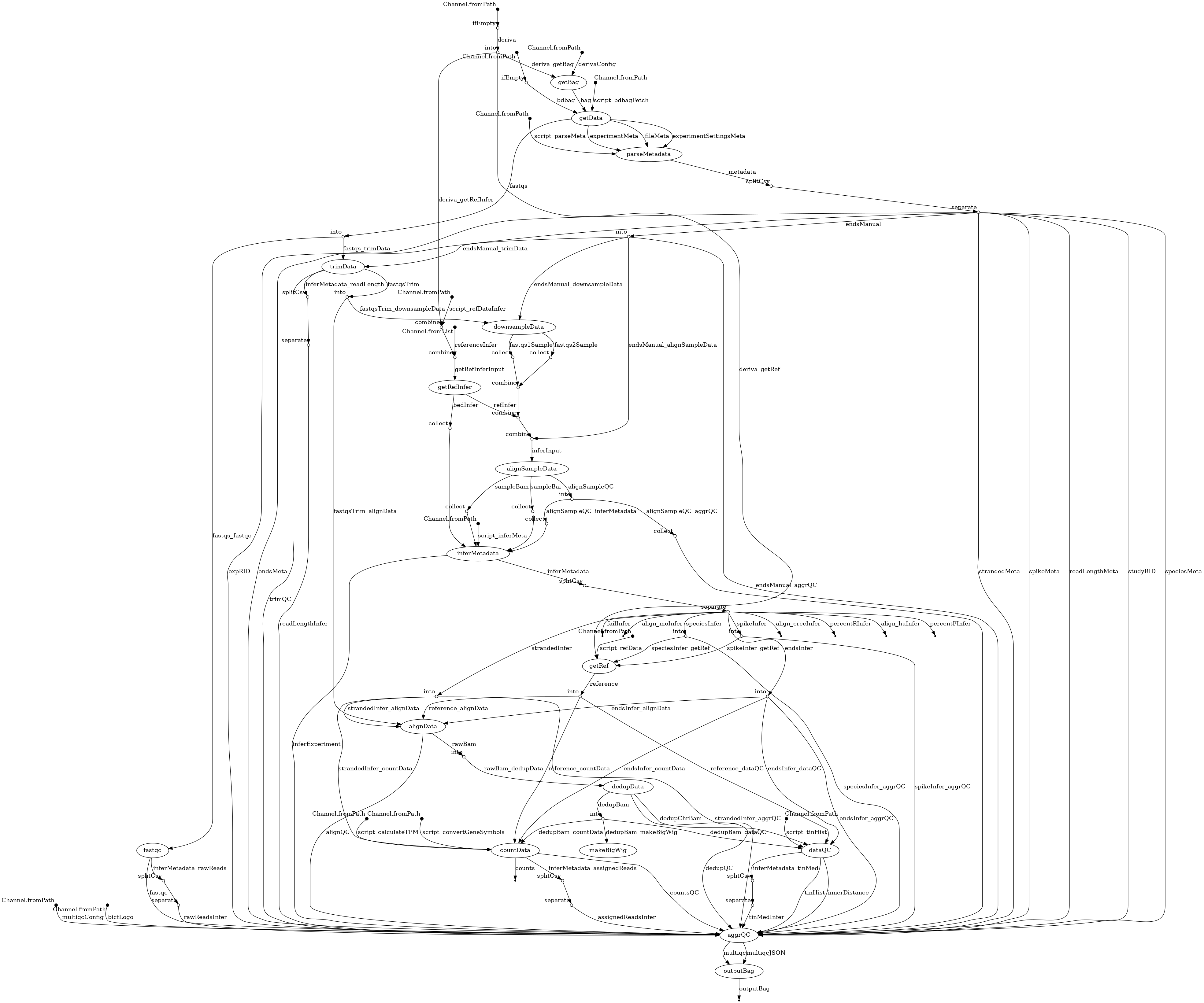
- docs/dag.png 0 additions, 0 deletionsdocs/dag.png
- workflow/nextflow.config 2 additions, 1 deletionworkflow/nextflow.config
- workflow/rna-seq.nf 59 additions, 34 deletionsworkflow/rna-seq.nf
docs/dag.png
100755 → 100644
| W: | H:
| W: | H: