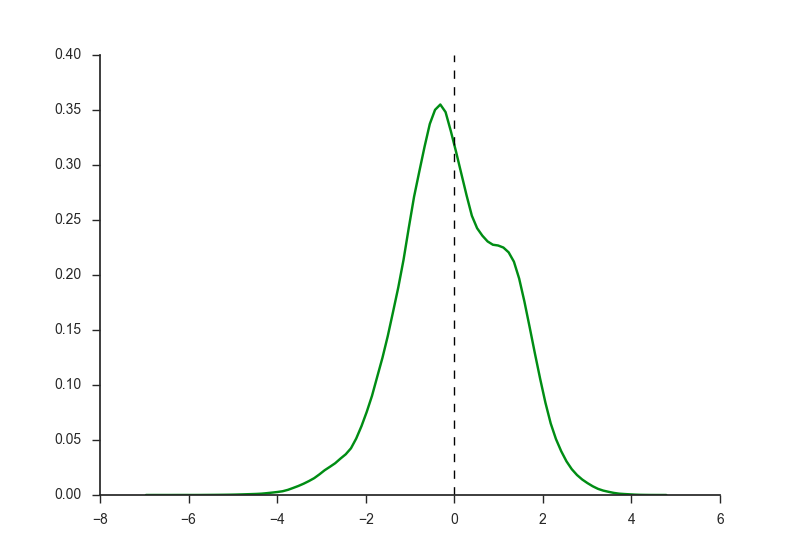
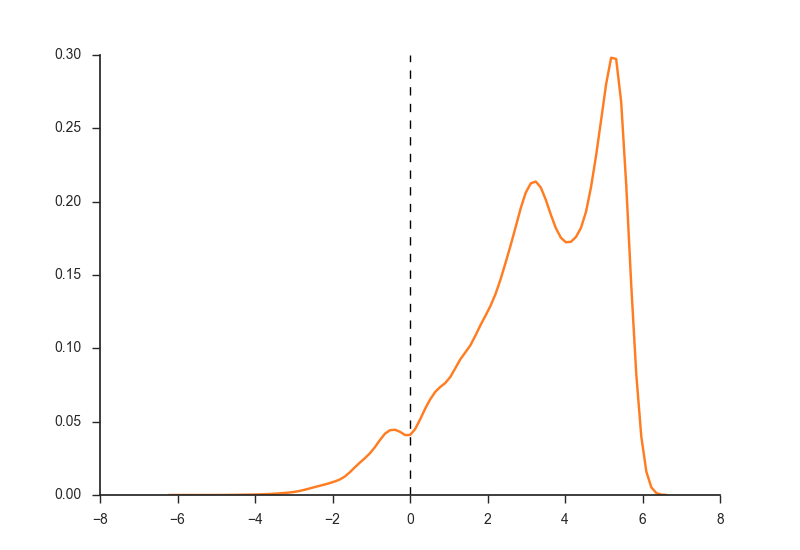
Show cutoffs for H3K4me3 and H3K27ac marks.
Showing
- H3K27ac_distribution.png 0 additions, 0 deletionsH3K27ac_distribution.png
- H3K4me3_distribution.png 0 additions, 0 deletionsH3K4me3_distribution.png
- cutoff_analysis.py 60 additions, 0 deletionscutoff_analysis.py
- h3k27ac_processing.sh 6 additions, 0 deletionsh3k27ac_processing.sh
- h3k4me3_processing.sh 4 additions, 0 deletionsh3k4me3_processing.sh
H3K27ac_distribution.png
0 → 100644
26.8 KiB
H3K4me3_distribution.png
0 → 100644
24.5 KiB
cutoff_analysis.py
0 → 100755