Showing
- analysis/GRO_seq_TFSEE/box_plot_cluster_1_tfs_fpkm.png 0 additions, 0 deletionsanalysis/GRO_seq_TFSEE/box_plot_cluster_1_tfs_fpkm.png
- analysis/GRO_seq_TFSEE/box_plot_cluster_2.png 0 additions, 0 deletionsanalysis/GRO_seq_TFSEE/box_plot_cluster_2.png
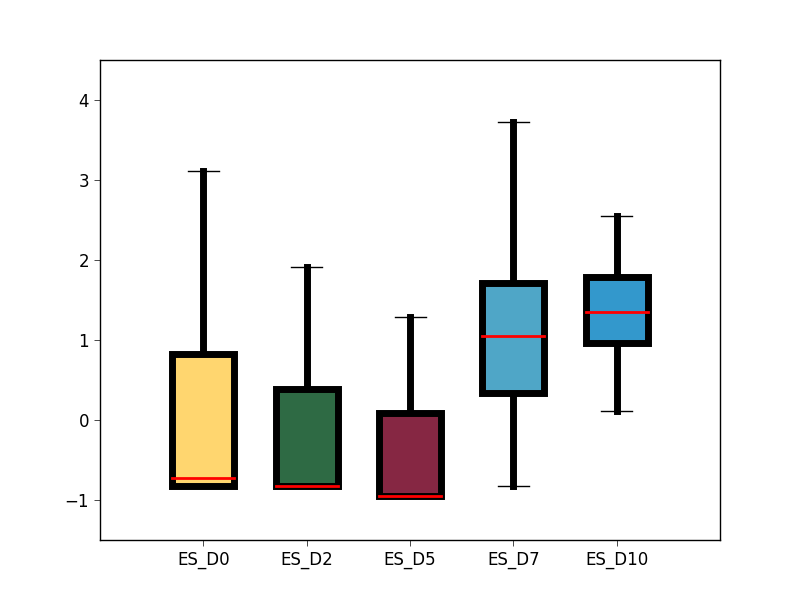
- analysis/GRO_seq_TFSEE/box_plot_cluster_2_enhancers_rpkm.png 0 additions, 0 deletionsanalysis/GRO_seq_TFSEE/box_plot_cluster_2_enhancers_rpkm.png
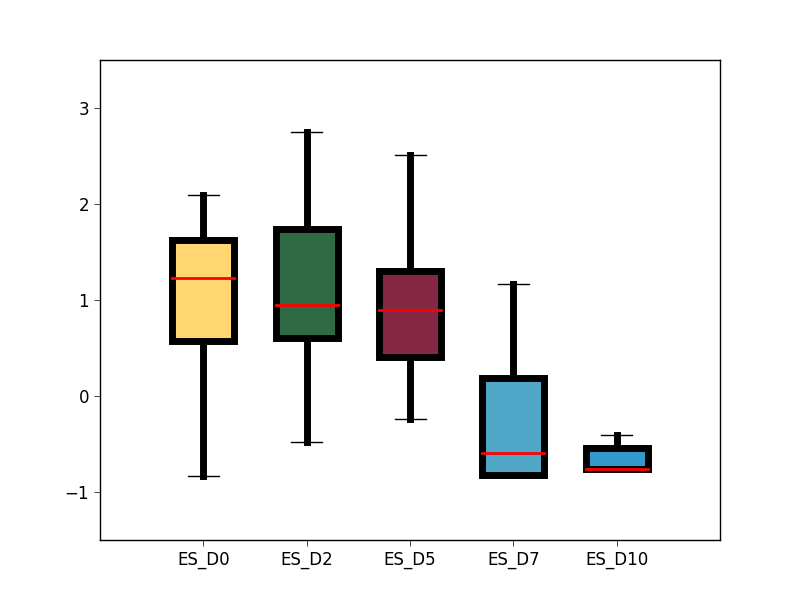
- analysis/GRO_seq_TFSEE/box_plot_cluster_2_genes_fpkm.png 0 additions, 0 deletionsanalysis/GRO_seq_TFSEE/box_plot_cluster_2_genes_fpkm.png
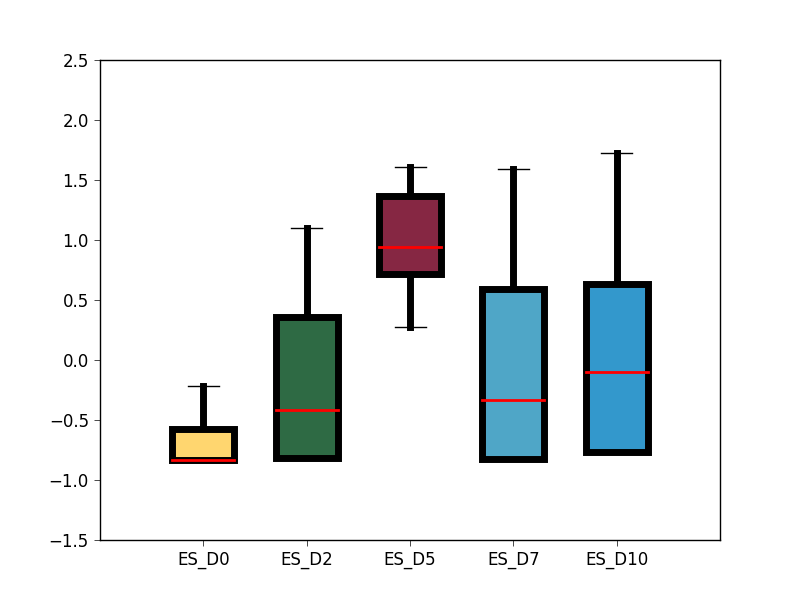
- analysis/GRO_seq_TFSEE/box_plot_cluster_2_tfs_fpkm.png 0 additions, 0 deletionsanalysis/GRO_seq_TFSEE/box_plot_cluster_2_tfs_fpkm.png
- analysis/GRO_seq_TFSEE/box_plot_cluster_3.png 0 additions, 0 deletionsanalysis/GRO_seq_TFSEE/box_plot_cluster_3.png
- analysis/GRO_seq_TFSEE/box_plot_cluster_3_enhancers_rpkm.png 0 additions, 0 deletionsanalysis/GRO_seq_TFSEE/box_plot_cluster_3_enhancers_rpkm.png
- analysis/GRO_seq_TFSEE/box_plot_cluster_3_genes_fpkm.png 0 additions, 0 deletionsanalysis/GRO_seq_TFSEE/box_plot_cluster_3_genes_fpkm.png
- analysis/GRO_seq_TFSEE/box_plot_cluster_3_tfs_fpkm.png 0 additions, 0 deletionsanalysis/GRO_seq_TFSEE/box_plot_cluster_3_tfs_fpkm.png
- analysis/GRO_seq_TFSEE/box_plot_cluster_4.png 0 additions, 0 deletionsanalysis/GRO_seq_TFSEE/box_plot_cluster_4.png
- analysis/GRO_seq_TFSEE/box_plot_cluster_4_enhancers_rpkm.png 0 additions, 0 deletionsanalysis/GRO_seq_TFSEE/box_plot_cluster_4_enhancers_rpkm.png
- analysis/GRO_seq_TFSEE/box_plot_cluster_4_genes_fpkm.png 0 additions, 0 deletionsanalysis/GRO_seq_TFSEE/box_plot_cluster_4_genes_fpkm.png
- analysis/GRO_seq_TFSEE/box_plot_cluster_4_tfs_fpkm.png 0 additions, 0 deletionsanalysis/GRO_seq_TFSEE/box_plot_cluster_4_tfs_fpkm.png
- analysis/GRO_seq_TFSEE/closest_genes.py 226 additions, 0 deletionsanalysis/GRO_seq_TFSEE/closest_genes.py
- analysis/GRO_seq_TFSEE/closest_genes.sh 14 additions, 0 deletionsanalysis/GRO_seq_TFSEE/closest_genes.sh
- analysis/GRO_seq_TFSEE/cluster1_enriched_tfs.csv 65 additions, 0 deletionsanalysis/GRO_seq_TFSEE/cluster1_enriched_tfs.csv
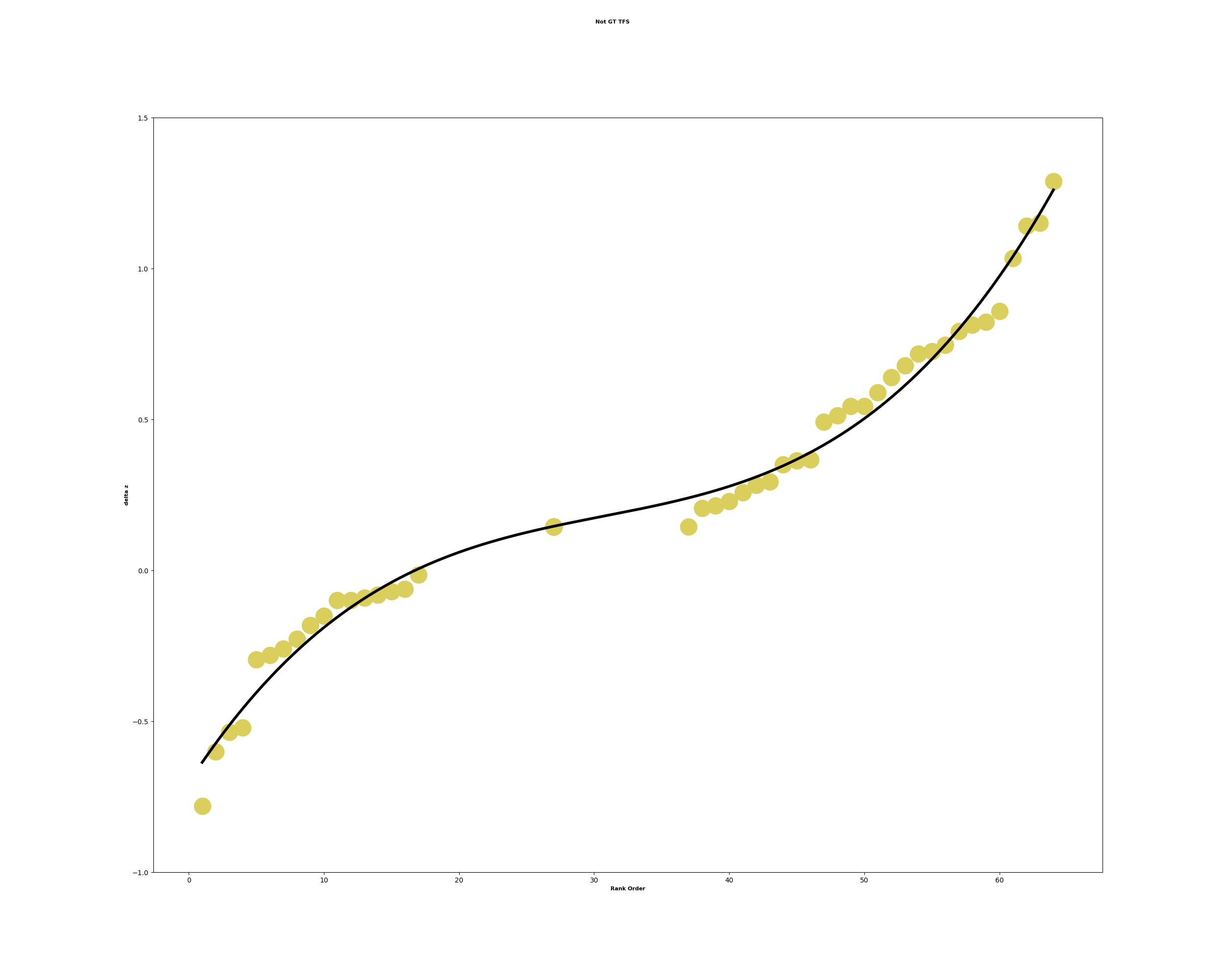
- analysis/GRO_seq_TFSEE/cluster1_enriched_tfs.png 0 additions, 0 deletionsanalysis/GRO_seq_TFSEE/cluster1_enriched_tfs.png
- analysis/GRO_seq_TFSEE/cluster2_enriched_tfs.csv 25 additions, 0 deletionsanalysis/GRO_seq_TFSEE/cluster2_enriched_tfs.csv
- analysis/GRO_seq_TFSEE/cluster2_enriched_tfs.png 0 additions, 0 deletionsanalysis/GRO_seq_TFSEE/cluster2_enriched_tfs.png
- analysis/GRO_seq_TFSEE/cluster3_enriched_tfs.csv 35 additions, 0 deletionsanalysis/GRO_seq_TFSEE/cluster3_enriched_tfs.csv
11.6 KiB
13.9 KiB
14.1 KiB
11.4 KiB
12 KiB
9.87 KiB
11.4 KiB
11.1 KiB
12 KiB
10.1 KiB
12.9 KiB
12 KiB
12.6 KiB
analysis/GRO_seq_TFSEE/closest_genes.py
0 → 100644
analysis/GRO_seq_TFSEE/closest_genes.sh
0 → 100644
This diff is collapsed.
96.2 KiB
This diff is collapsed.
85.6 KiB
This diff is collapsed.