# Astrocyte RNASeq Workflow Package
## Workflow SOP
This SOP describes the analysis pipeline of RNA sequencing data. This pipeline includes (1) quality control, (2) variant calling analysis, (3) identification of fusion genes, and (4) statistical analyses of gene expression and isoform expression. The result R data of the statistical analysis can be visualized using a custom R shiny service.
For each file this workflow:
1) Trim the ends of sequences with remaining adapter or quality scores < 25. Remove any sequence less than 35bp after trimming, then generate a file for capturing information about how many sequences were trimmed.
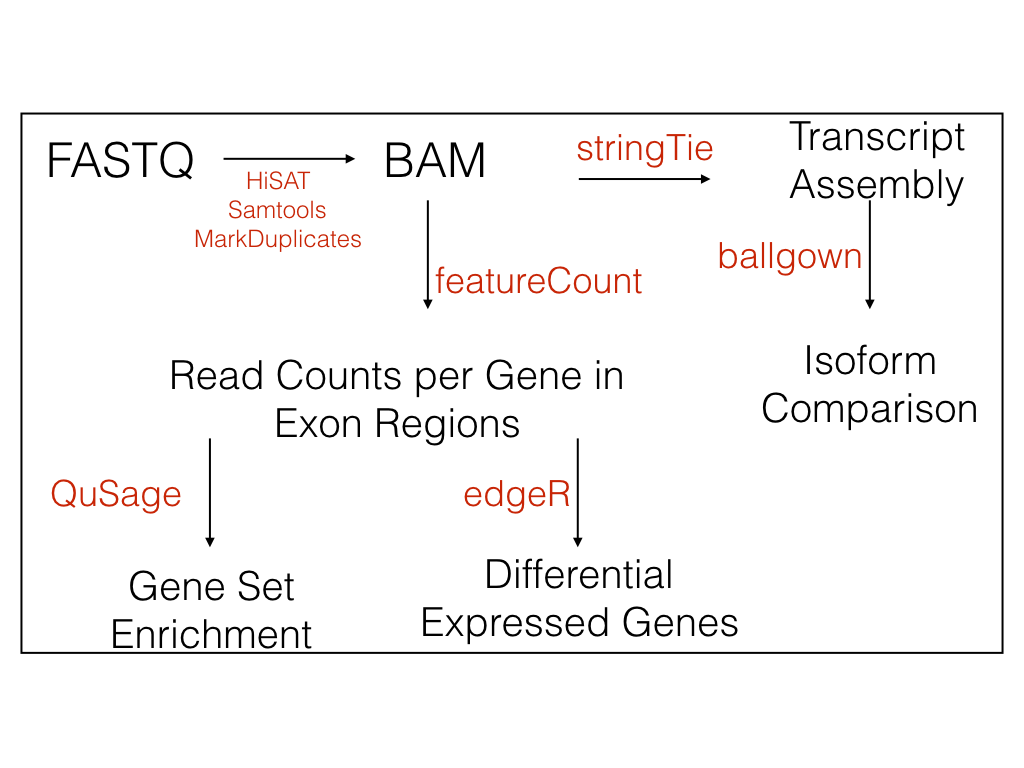
2) Trimmed Fastq files are aligned to the selected reference genome using HiSAT2 (Kim et al. 2015) or STAR (Dobin et al. 2012)
3) Marks Duplicates using SAMBAMBA
4) Features (genes, transcripts and exons) are counted using featureCounts (Liao et al 2014) and StringTie (Pertea et al. 2015) using the Gencode feature table(Harrow et al. 2012)
5) Basic pairwise differential expression analysis is performed using EdgeR (Robinson et al. 2010) and DESeq
6) Abundances of transcripts are calculated using ballgown (Frazee et al. 2014)
7) Identify gene fusions or fused transcripts using STAR-Fusion.
8) Identify coding variants:
a) Using GATK, repair the format problem of input BAM file, (2) add read group information, (3) handle N-split mapping of reads caused by RNA splicing, (4) base quality score recalibration (BQSR): detect systematic errors made by the sequencer when it estimates the quality score of each base call and build a model of covariation based on the data and a set of known variants, then adjust the base quality scores in the data based on the model.
b) Detect variants using SAMTOOLS with mapping quality >= 50 and base calling quality >= 20.
c) Detect low frequency variants using SAMTOOLS with variant allele fraction > 0.01 and depth > 50 and has been previously identified.
d) Detect variants using SpeedSeq
e) Detect variants using GATK
## Workflow Parameters
fastq - Choose one or more RNASeq read files to process.
genome - Choose a genomic reference (genome).
stranded - If a stranded library is created, please select the appropriate protocol to ensure strand specific analysis
markdups - The default is to remove duplicates, you can skip this function.
pairs - Indicate if this is paired-end or single-end sequencing data
dea - Perform Differential Expression analysis, please skip if there are < 2 sample groups
geneset - Select a set of known genesets for pathway analysis
fusion - Perform Gene Fusion Identification
variant - Perform variant detection
tumor - Perform Cancer specific analysis in variant detection
design - This file matches the fastq files to data abot the sample
The following columns are necessary, must be named as in template and can be in any order:
SampleID
This ID should match the name in the fastq file ie S0001.R1.fastq.gz the sample ID is S0001
Notes: SampleID shouldn't start with numbers ie 10C should be changed to S10C
SampleID should contain only alphanumeric characters ie the '_', '.', and '-' characters should be removed from "treated_sample.1-1"
SampleName
This ID can be the identifier of the researcher or clinician
SubjectID
Used in order to link samples from the same patient
SampleGroup
This is the group that will be used for pairwise differential expression analysis
FqR1
Name of the fastq file R1
FqR2
Name of the fastq file R2
There are some optional columns that might help with the analysis:
Tissue
Gender
CultureDate
SequenceRun
Organism
CellPopulation
Treatment
GeneticFeature (WT or KO)
Race
Ethnicity
Age
### Test Data
### Credits
This example worklow is derived from original scripts kindly contributed by the Bioinformatic Core Facility (BICF), Department of Bioinformatics
### Workflow SOP
### References
* FastQC: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ o http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/
* STAR-Fusion: https://github.com/STAR-Fusion/STAR-Fusion
* STAR: https://github.com/alexdobin/STAR
* Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013 Jan 1;29(1):15-21. doi: 10.1093/bioinformatics/bts635. Epub 2012 Oct 25. PubMed PMID: 23104886; PubMed Central PMCID: PMC3530905.
* HISAT, StringTie and Ballgown protocol
* Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016 Sep;11(9):1650-67.
* StringTie: https://ccb.jhu.edu/software/stringtie/
* Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015 Mar;33(3):290-5.
* Ballgown
* http://biorxiv.org/content/biorxiv/early/2014/09/05/003665.full.pdf o http://bioconductor.org/packages/release/bioc/html/ballgown.html o https://github.com/alyssafrazee/ballgown
* HISAT: https://ccb.jhu.edu/software/hisat2/index.shtml
* Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015 Apr;12(4):357-60.
* SAMtools: http://samtools.sourceforge.net/
* Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009 Aug 15;25(16):2078-9.
* Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011 Nov 1;27(21):2987-93.
* Sambamba: http://lomereiter.github.io/sambamba/
* Tarasov A, Vilella AJ, Cuppen E, Nijman IJ, Prins P. Sambamba: fast processing of NGS alignment formats. Bioinformatics. 2015 Jun 15;31(12):2032-4. * Picard: https://broadinstitute.github.io/picard/
* SpeedSeq: https://github.com/hall-lab/speedseq
* Chiang C, Layer RM, Faust GG, Lindberg MR, Rose DB, Garrison EP, Marth GT, Quinlan AR, Hall IM. SpeedSeq: ultra-fast personal genome analysis and interpretation. Nat Methods. 2015 Oct;12(10):966-8.
* GATK: https://software.broadinstitute.org/gatk/
* McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010 Sep;20(9):1297-303.
* DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011 May;43(5):491-8.
* Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, Banks E, Garimella KV, Altshuler D, Gabriel S, DePristo MA. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11.10.1-33. o http://gatkforums.broadinstitute.org/gatk * COSMIC: http://cancer.sanger.ac.uk/cosmic
* BEDTools: http://bedtools.readthedocs.io/en/latest/
* Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010 Mar 15;26(6):841-2. * VCFtools: http://vcftools.sourceforge.net/index.html
* Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R; 1000 Genomes Project Analysis Group. The variant call format and VCFtools. Bioinformatics. 2011 Aug 1;27(15):2156-8.
* SnpSift: http://snpeff.sourceforge.net/
* Cingolani P, Patel VM, Coon M, Nguyen T, Land SJ, Ruden DM, Lu X. Using Drosophila melanogaster as a Model for Genotoxic Chemical Mutational Studies with a New Program, SnpSift. Front Genet. 2012 Mar 15;3:35.
* featureCounts: http://subread.sourceforge.net
* Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014 Apr 1;30(7):923-30.
* edgeR: https://bioconductor.org/packages/release/bioc/html/edgeR.html
* Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010 Jan 1;26(1):139-40.
* McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012 May;40(10):4288-97.